This is the R implementation of the Clustering-Informed Shared-Structure Variational Autoencoder (CISS-VAE) for imputation. The Python implementation can be found here and is available for installation on PyPI.
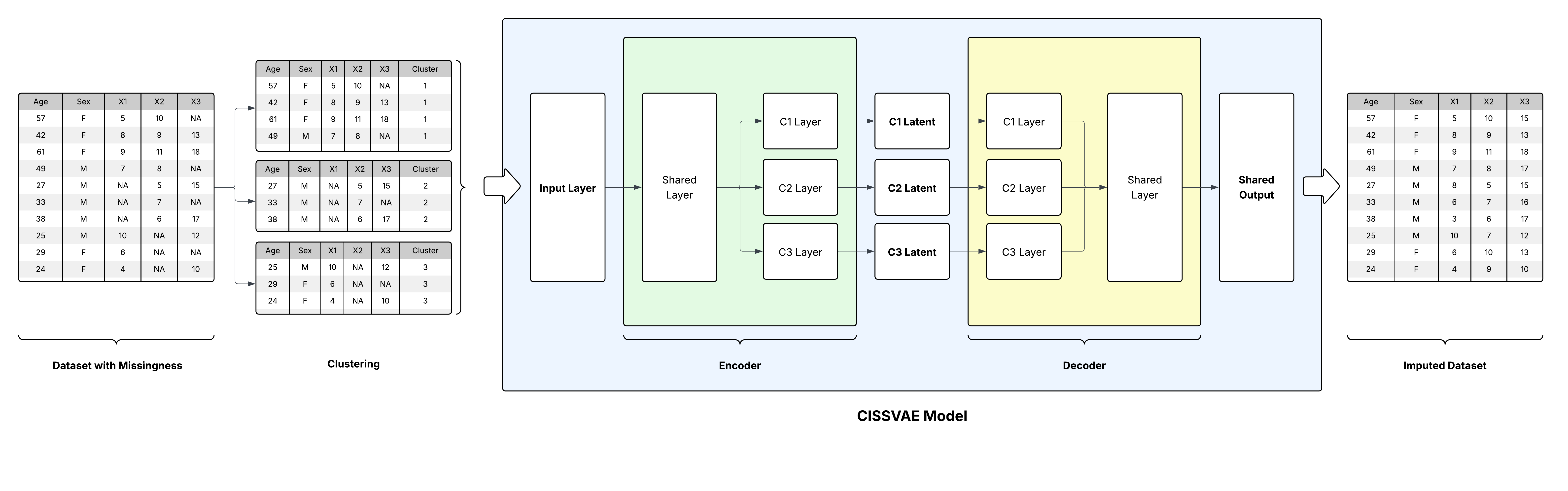
CISS-VAE is a flexible deep learning model for missing data imputation that accommodates all three types of missing data mechanisms: Missing Completely At Random (MCAR), Missing At Random (MAR), and Missing Not At Random (MNAR). While it is particularly well-suited to MNAR scenarios where missingness patterns carry informative signals, CISS-VAE also functions effectively under MAR assumptions. For more information, please check out our publication!
Click Here for More Information
A key feature of CISS-VAE is the use of unsupervised clustering to capture distinct patterns of missingness. Alongside cluster-specific representations, the method leverages shared encoder and decoder layers. This allows for knowledge transfer across clusters and enhances parameter stability, which is especially important when some clusters have small sample sizes. In situations where the data do not naturally partition into meaningful clusters, the model defaults to a pooled representation, preventing unnecessary complications from cluster-specific components.
Installation
rCISSVAE can now be installed via CRAN:
install.packages("rCISSVAE")For the latest development version, please install via github:
Install devtools or remotes if not already installed:
install.packages("remotes")
# or
install.packages("devtools")The rCISSVAE package can then be installed with:
remotes::install_github("CISS-VAE/rCISS-VAE")
# or
devtools::install_github("CISS-VAE/rCISS-VAE")Ensuring correct virtual environment for reticulate
This package uses reticulate to interface with the python version of the package ciss-vae, which can be installed via pip.
If you are comfortable creating a venv or conda environment and installing the package, great! Then all you need to do is tell reticulate where to point.
For Venv
reticulate::use_virtualenv("./.venv", required = TRUE)For conda
reticulate::use_condaenv("myenv", required = TRUE)Virtual environment helper function
If you do not want to manually create the virtual environment, you can use the helper function create_cissvae_env() to create a virtual environment (venv).
create_cissvae_env(
envname = "cissvae_environment", ## name of environment
path = NULL, ## add path to wherever you want virtual environment to be. If null will create environment in current working directory
install_python = FALSE, ## set to TRUE if you want create_cisssvae_env to install python for you
python_version = "3.10" ## set to whatever version you want >=3.10. Python 3.10 or 3.11 recommended
)Once the environment is created, activate it using:
reticulate::use_virtualenv("./cissvae_environment", required = TRUE)
# In other words,
# reticulate::use_virtualenv("./your_environment_name", required = TRUE)Quickstart
The input dataset should be a matrix or data.frame and missing values should be represented with NA or NaN. Precomputed clusters should be passed as a numeric vector of length ‘n’ for an ‘n x p’ dataset. For more information on having run_cissvae() handle the clustering see the ‘Clustering Features by Missingness Patterns’ vignette.
library(reticulate)
library(rCISSVAE)
data(df_missing)
data(clusters)
dat = run_cissvae(
data = df_missing,
index_col = "index",
val_proportion = 0.1, ## pass a vector for different proportions by cluster
columns_ignore = c("Age", "Salary", "ZipCode10001", "ZipCode20002", "ZipCode30003"), ## If there are columns in addition to the index you want to ignore when selecting validation set, list them here. In this case, we ignore the 'demographic' columns because we do not want to remove data from them for validation purposes.
clusters = clusters$clusters, ## we have precomputed cluster labels so we pass them here
epochs = 5,
return_silhouettes = FALSE,
return_history = TRUE, # Get detailed training history
verbose = FALSE,
return_model = TRUE, ## Allows for plotting model schematic
device = "cpu", # Explicit device selection
layer_order_enc = c("unshared", "shared", "unshared"),
layer_order_dec = c("shared", "unshared", "shared"),
latent_shared = FALSE,
output_shared = FALSE
)
plot_vae_architecture(model = dat$model)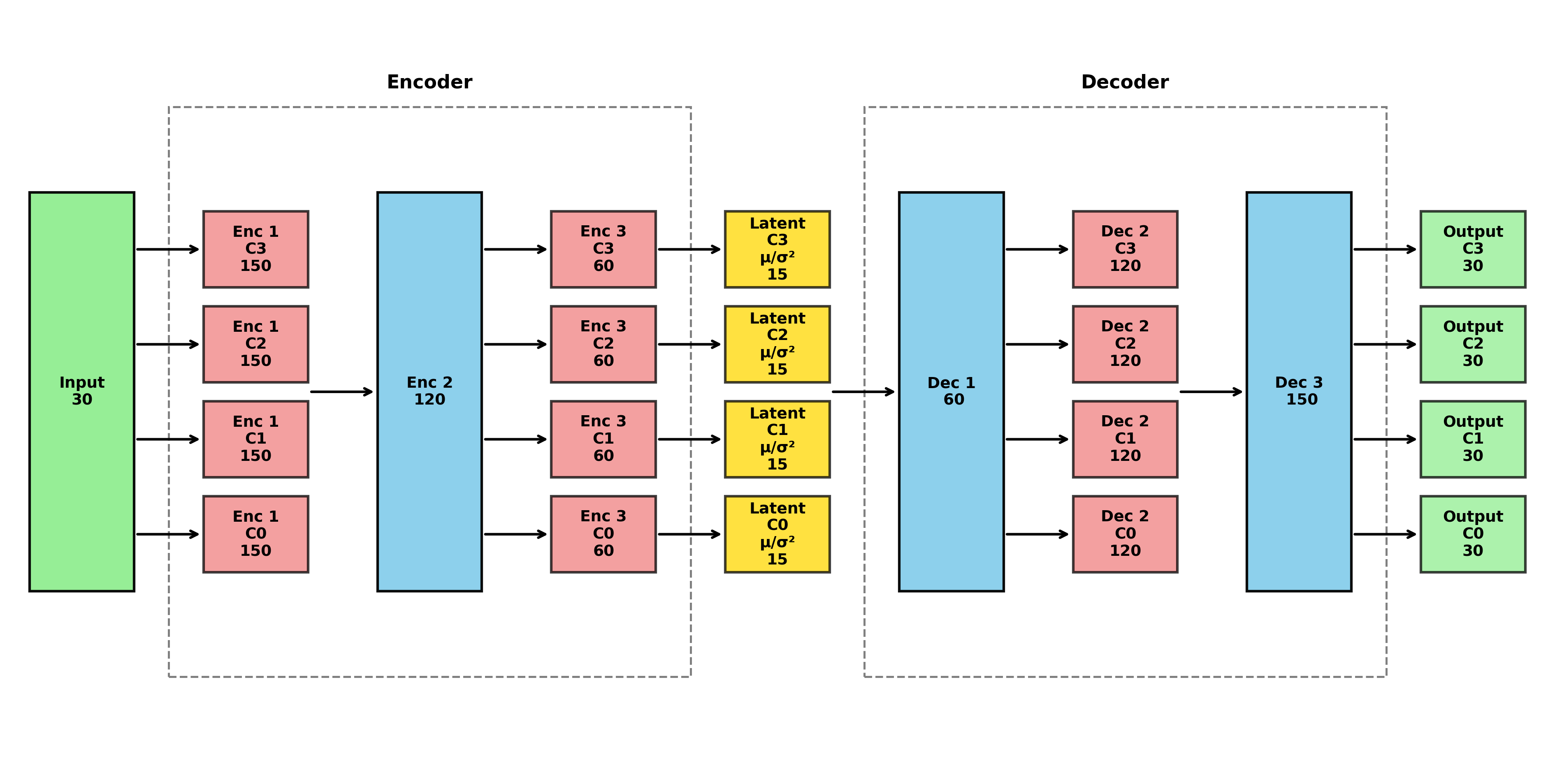
For a full tutorial, see vignette.
Citation
If you use our package in your research, please consider citing our recent publication!
Y. Khadem Charvadeh, K. Seier, K. S. Panageas, D. Vaithilingam, M.Gönen, and Y. Chen, “Clustering-Informed Shared-Structure Variational Autoencoder for Missing Data Imputation in Large-Scale Healthcare Data,” Statistics in Medicine 44, no. 28-30 (2025): e70335, https://doi.org/10.1002/sim.70335.
Authors
- Yasin Khadem Charvadeh
- Kenneth Seier
- Katherine S. Panageas
- Danielle Vaithilingam
- Mithat Gönen
- Yuan Chen (corresponding author) — cheny19@mskcc.org